Rapid cloud setup and research integrity with no platform lock-in
Code Ocean is for academic labs that want a reproducible, cost-effective way to work in the cloud without time-consuming setup or DevOps overhead. Free for labs of up to 25.












Key benefits for universities
Reduce and control cloud compute costs
Idleness detection automatically shuts off compute resources after an admin-defined period. Administrators can view costs as a whole or get cost per run, dataset, user, or project, and they can optionally limit the availability of dedicated machines.

Setup new researchers in the cloud in minutes
Individual researchers can build their own environments with Dockerfile autogeneration, or use a fully-loaded environment provided at the organization level. Compute resources are then selected via popup menu.

Improve reproducibility and research integrity
Compute Capsules are a shareable, traceable, reproducible encapsulation of the code, data, and environments used in computational research, version controlled and linked to the results they produce.

Export to run on HPC, maintain result lineage
Capsules and Pipelines can be run in the cloud, but can also be exported to run on local HPCs, then brought back into the Code Ocean system to maintain full result lineage and provenance with the Lineage Graph tool.

Build a library of your computational research
All computational work is tracked and recorded in the Code Ocean system, with controls available to transfer assets to others if needed. Collections help make work visible to other users and groups.

No platform lock-in: export anything, at any time
Code Ocean is built on open-source technology and designed to make analytics FAIRR: findable, accessible, interoperable, reusable, and reproducible. Export any Capsule or Pipeline at any time to run outside of the platform or transfer elsewhere.

Research integrity for journal code submission
Connect to the Open Science Library (OSL): the industry standard for computational scientific reproducibility and research integrity in peer-reviewed journals.

Who uses Code Ocean?
“Code Ocean totally solves tracking and reproducing analysis for researchers and increases trust in the results.”

Benjamin Haibe-Kains, Ph.D
Director of In Silico Biology, Ochre Bio
“Code Ocean sped up our internal image pre-processing computational workflow by at least 10x, improving collaboration and productivity between our global teams ... removing the need for painful cloud computing infrastructure set up.”

Dimitris Polychronopoulos
Director of In Silico Biology, Ochre Bio
“Code Ocean's self-service capabilities make it easy for our scientists to do their work reproducibly. New users to the platform can get far with just a little support, giving our engineers time to focus on domain-specific challenges.”

Dr. David Feng
Director of Scientific Computing, Allen Institute





Key product features

Built for Computational Science
-
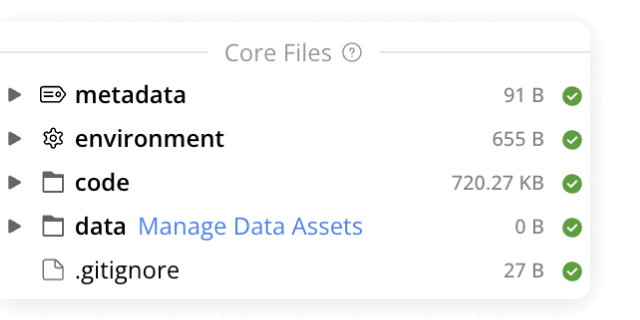
Data analysis

Data analysis
Use ready-made template Compute Capsules to analyze your data, develop your data analysis workflow in your preferred language and IDE using any open-source software, and take advantage of built-in containerization to guarantee reproducibility.
-
Data management

Data management
Manage your organization's data and control who has access to it. Built specifically to meet all FAIR principles, data management in Code Ocean uses custom metadata and controlled vocabularies to ensure consistency and improve searchability.
-
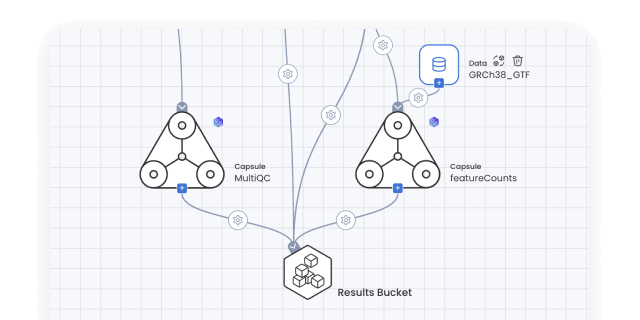
Bioinformatics pipelines

Bioinformatics pipelines
Build, configure and monitor bioinformatics pipelines from scratch using a visual builder for easy set-up. Or, import from nf-core in one click for instant access to a curated set of best practice analysis pipelines. Runs on AWS Batch out-of-the-box, so your pipelines scale automatically. No setup needed.
-
ML models
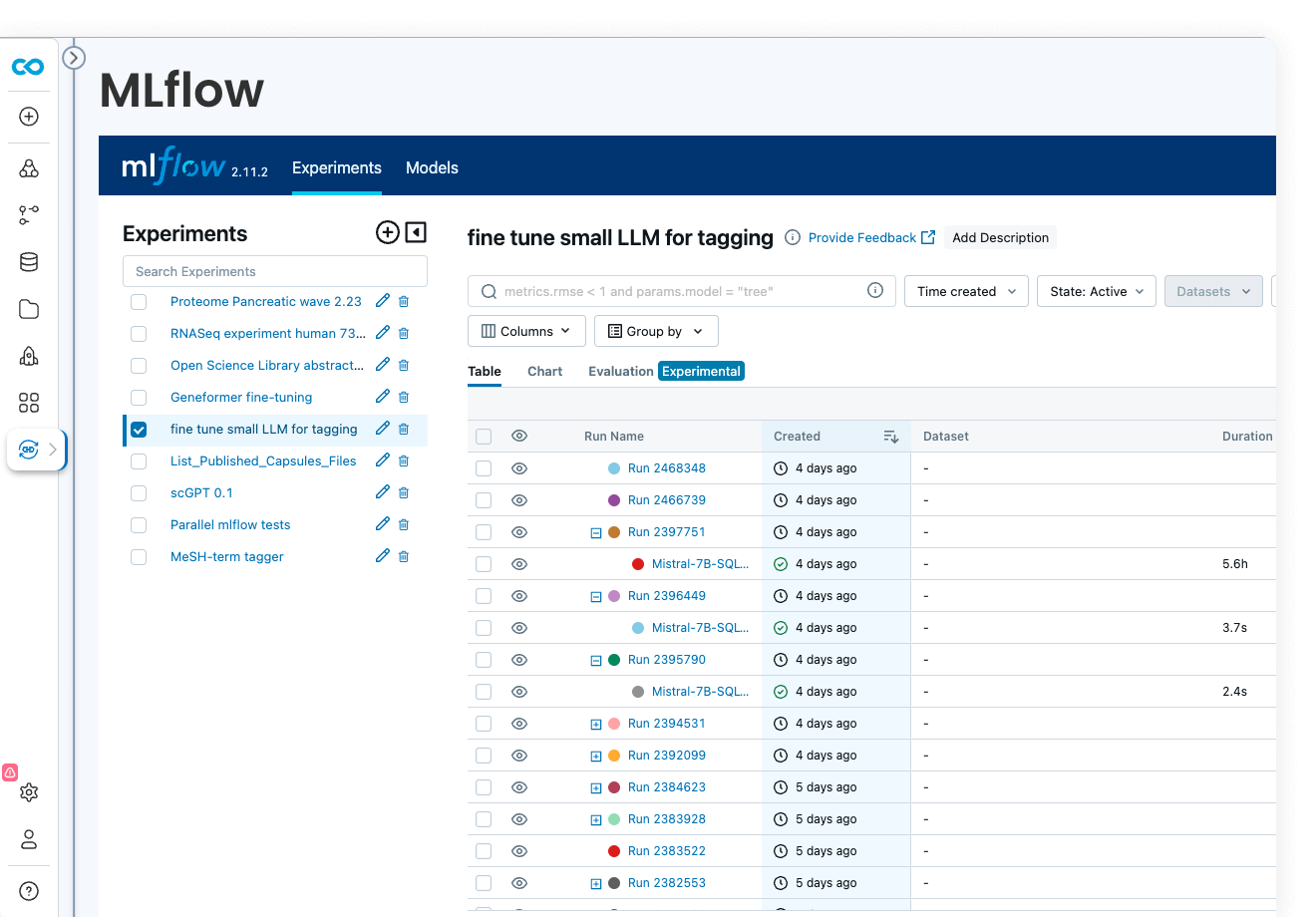

ML model development
Code Ocean is uniquely suited for Artificial Intelligence, Machine Learning, Deep Learning, and Generative AI. Install GPU-ready environments and provision GPU resources in a few clicks. Integration with MLFlow allows you to develop models, track parameters, manage models from development to production, while enjoying out-of-the-box reproducibility and lineage.
-
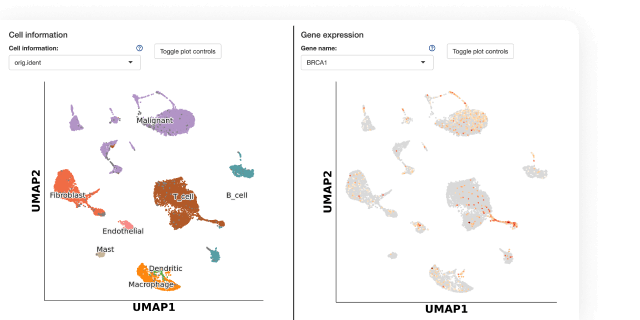
Multiomics


Multiomics
Analyze and work with large multimodal datasets efficiently using scalable compute and storage resources, cached packages for R and Python, preloaded multiomics analysis software that works out of the box and full lineage and reproducibility.
-
Imaging
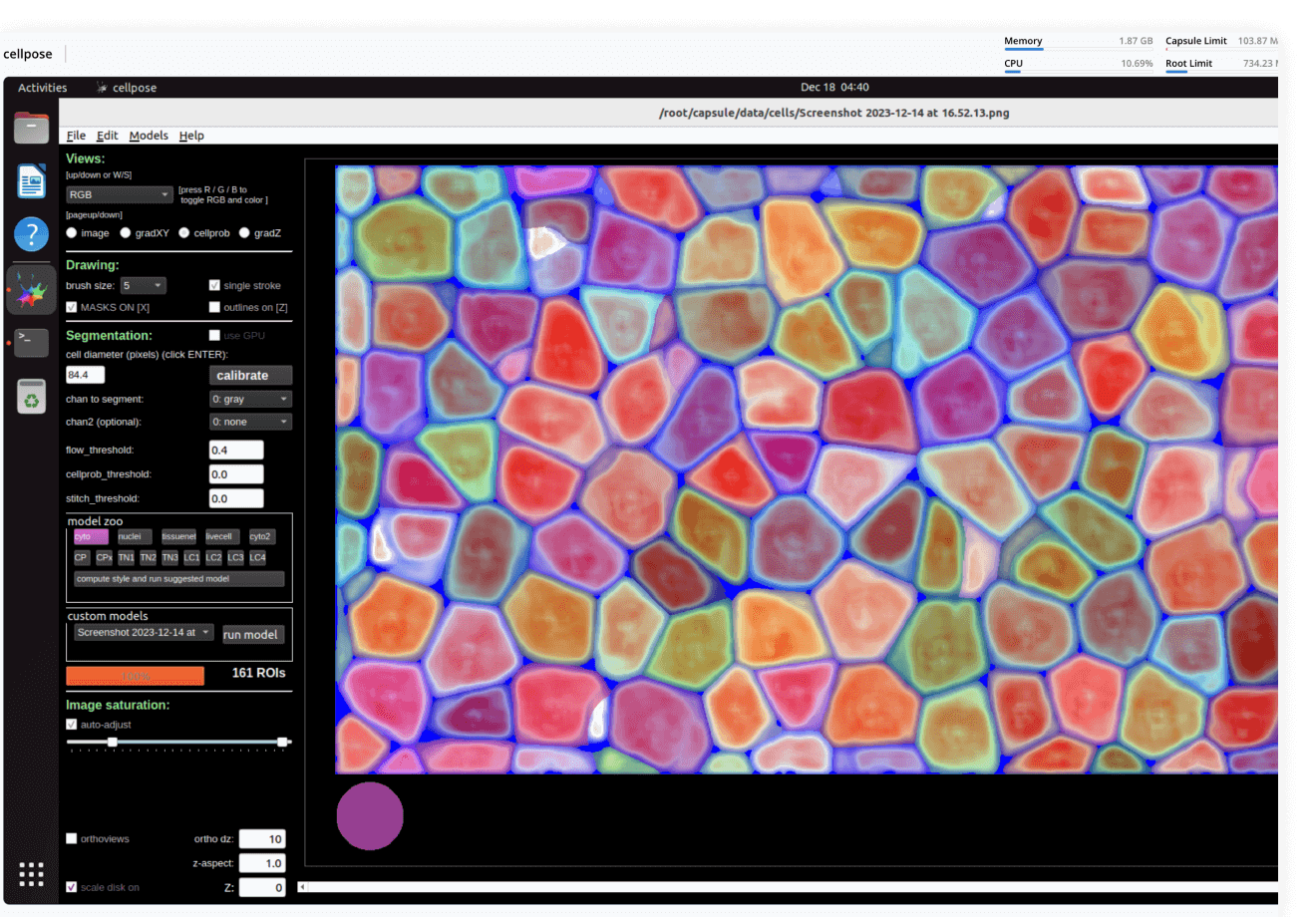

Imaging
Process images using a variety of tools: from dedicated desktop applications to custom-written deep learning pipelines, from a few individual files to petabyte-sized datasets. No DevOps required, always with lineage.
-
Cloud management


Cloud management
Code Ocean makes it easy to manage data and provision compute: CPUs, GPUs, and RAM. Assign flex machines and dedicated machines to manage what is available to your users. Spot instances, idleness detection, and automated shutdown help reduce cloud costs.
-
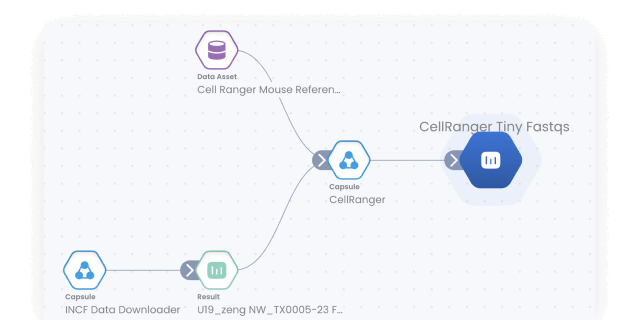
Data/model provenance

Data/model provenance
Keep track of all data and results with automated result provenance and lineage graph generation. Assess reproducibility with a visual representation of every Capsule, Pipeline, and Data asset involved in a computation.

Data analysis
Use ready-made template Compute Capsules to analyze your data, develop your data analysis workflow in your preferred language and IDE using any open-source software, and take advantage of built-in containerization to guarantee reproducibility.

Data management
Manage your organization's data and control who has access to it. Built specifically to meet all FAIR principles, data management in Code Ocean uses custom metadata and controlled vocabularies to ensure consistency and improve searchability.

Bioinformatics pipelines
Build, configure and monitor bioinformatics pipelines from scratch using a visual builder for easy set-up. Or, import from nf-core in one click for instant access to a curated set of best practice analysis pipelines. Runs on AWS Batch out-of-the-box, so your pipelines scale automatically. No setup needed.

ML model development
Code Ocean is uniquely suited for Artificial Intelligence, Machine Learning, Deep Learning, and Generative AI. Install GPU-ready environments and provision GPU resources in a few clicks. Integration with MLFlow allows you to develop models, track parameters, manage models from development to production, while enjoying out-of-the-box reproducibility and lineage.

Multiomics
Analyze and work with large multimodal datasets efficiently using scalable compute and storage resources, cached packages for R and Python, preloaded multiomics analysis software that works out of the box and full lineage and reproducibility.

Imaging
Process images using a variety of tools: from dedicated desktop applications to custom-written deep learning pipelines, from a few individual files to petabyte-sized datasets. No DevOps required, always with lineage.

Cloud management
Code Ocean makes it easy to manage data and provision compute: CPUs, GPUs, and RAM. Assign flex machines and dedicated machines to manage what is available to your users. Spot instances, idleness detection, and automated shutdown help reduce cloud costs.

Data/model provenance
Keep track of all data and results with automated result provenance and lineage graph generation. Assess reproducibility with a visual representation of every Capsule, Pipeline, and Data asset involved in a computation.

.png?length=720&name=September%20Webinar%20-%20On%20Demand%20(1).png)
