Pipelines
Connect, automate, parallelize, and scale computational work. Build with the visual editor and auto-generate Nextflow code, import from nf-core, or write your own.
Key capabilities
Build visually with Nextflow auto-generation
Build pipelines in a visual editor while Code Ocean writes Nextflow in the background in real time. Import pre-existing pipelines directly from nf-core or clone from a Git repo. Advanced users can unlock and write their own Nextflow script.

Import pipelines directly from nf-core
Import ready-made, community-created bioinformatics pipelines from nf-core and use them directly in Code Ocean. No further setup or config needed; just define your parameters and run in the cloud.

Define compute resources for every step
Define CPU, GPU and RAM requirements for every step of the pipeline, adjusting for computational needs or other constraints.

Parallelize and scale with AWS Batch
Code Ocean comes with a fully configured AWS Batch instance out-of-the-box. With zero set-up, Pipelines will run on AWS Batch to parallelize work and use EBS autoscaling to automatically adjust storage needs up and down as the Pipeline runs.

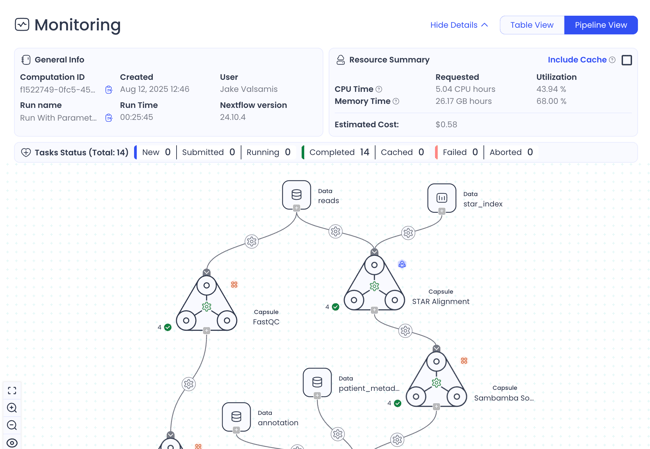
Monitor runs with real-time visibility and control
The monitoring dashboard provides run status, resource summary, processes overview, task details, logs, file preview, and more. Debug faster and ensure your Pipeline runs are cost-efficient.
Share Pipelines with other users or groups
Share Pipelines with other users or groups on your team with user permissions built-in. Work on the Pipeline collaboratively, or clone and adapt for your own needs. Option to share all assets associated.

How Pipelines work with the rest of the Code Ocean platform
Frequently asked questions
 Pipelines
Pipelines
What language are Pipelines based on?
Nextflow. Pipelines have native integration of nextflow, with nextflow scripts automatically created and written as uesrs work in the visual pipeline builder.
Which nextflow DSL is supported?
Both. Building Pipelines with Code Ocean’s Pipeline Builder UI uses DSL1. Custom pipelines support both DSL1 and DSL2.
Do we always have to use the visual pipeline builder tool?
No. Pipelines can be unlocked for users to write their own custom script in Nextflow. Users can fully leverage all Nextflow capabilities within a custom pipeline.
Can you assign different computing resources for different parts of the pipeline?
Yes. Each Capsule can have different computing resources applied (when part of a pipeline in the visual UI), depending on requirements.
I already have a pipeline I want to use. Can I bring that into Code Ocean?
Yes. Users can import an existing pipeline from a git repository, import from nf-core, or upload files to make their custom pipeline. This allows them to quickly create pipelines when first starting with Code Ocean.

How Ochre Bio turned a manual workflow into a pipeline
Built for Computational Science
-
Data analysis

Data analysis
Use ready-made template Compute Capsules to analyze your data, develop your data analysis workflow in your preferred language and IDE using any open-source software, and take advantage of built-in containerization to guarantee reproducibility.
-
Data management

Data management
Manage your organization's data and control who has access to it. Built specifically to meet all FAIR principles, data management in Code Ocean uses custom metadata and controlled vocabularies to ensure consistency and improve searchability.
-
Bioinformatics pipelines

Bioinformatics pipelines
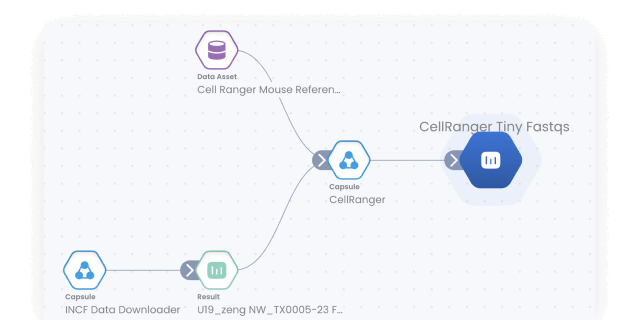
Build, configure and monitor bioinformatics pipelines from scratch using a visual builder for easy set-up. Or, import from nf-core in one click for instant access to a curated set of best practice analysis pipelines. Runs on AWS Batch out-of-the-box, so your pipelines scale automatically. No setup needed.
-
ML models

ML model development
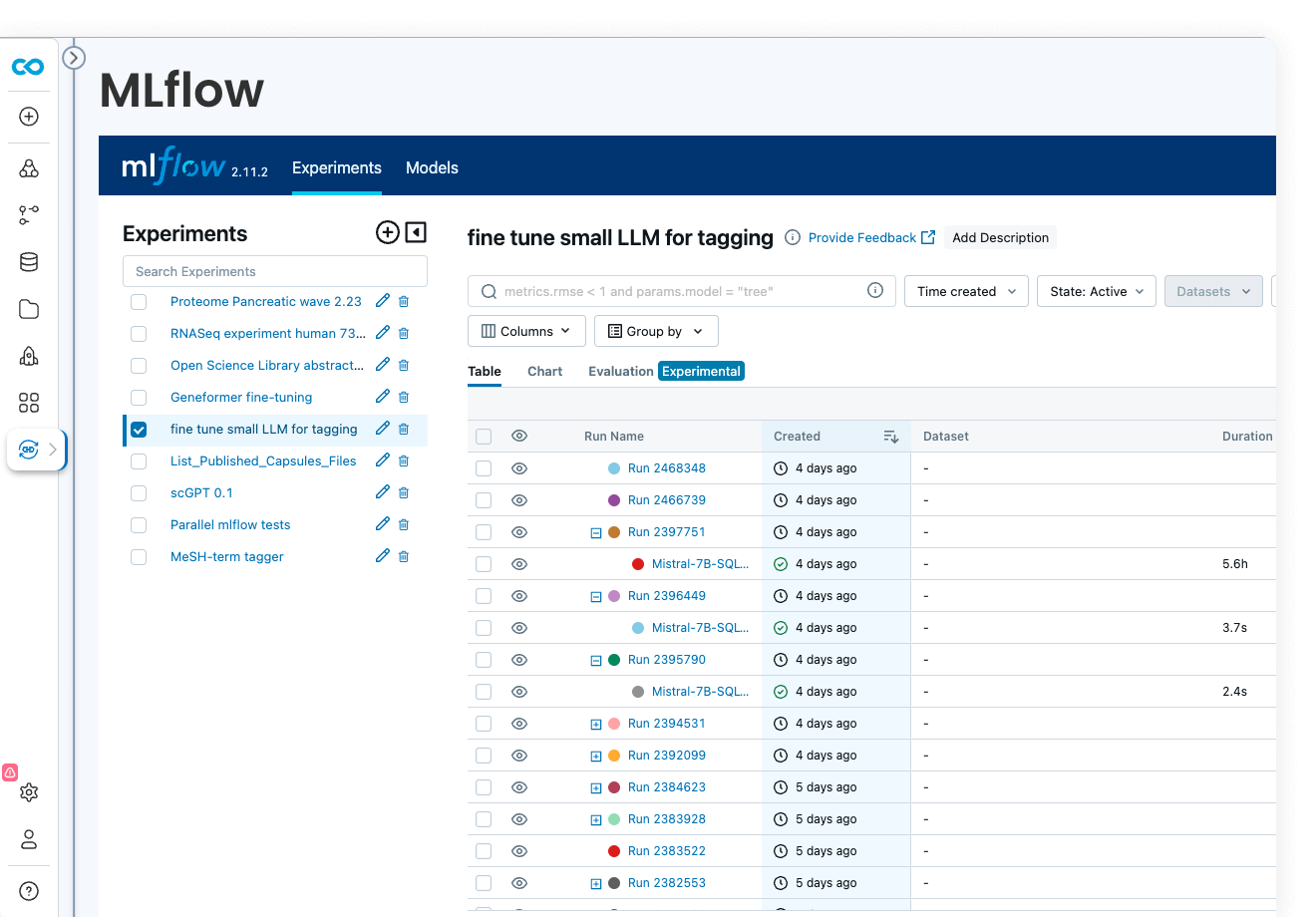
Code Ocean is uniquely suited for Artificial Intelligence, Machine Learning, Deep Learning, and Generative AI. Install GPU-ready environments and provision GPU resources in a few clicks. Integration with MLFlow allows you to develop models, track parameters, manage models from development to production, while enjoying out-of-the-box reproducibility and lineage.
-
Multiomics


Multiomics
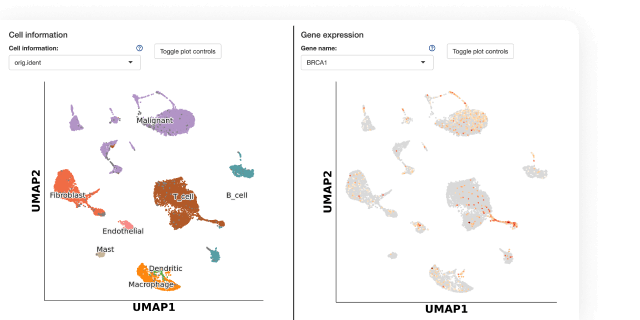
Analyze and work with large multimodal datasets efficiently using scalable compute and storage resources, cached packages for R and Python, preloaded multiomics analysis software that works out of the box and full lineage and reproducibility.
-
Imaging
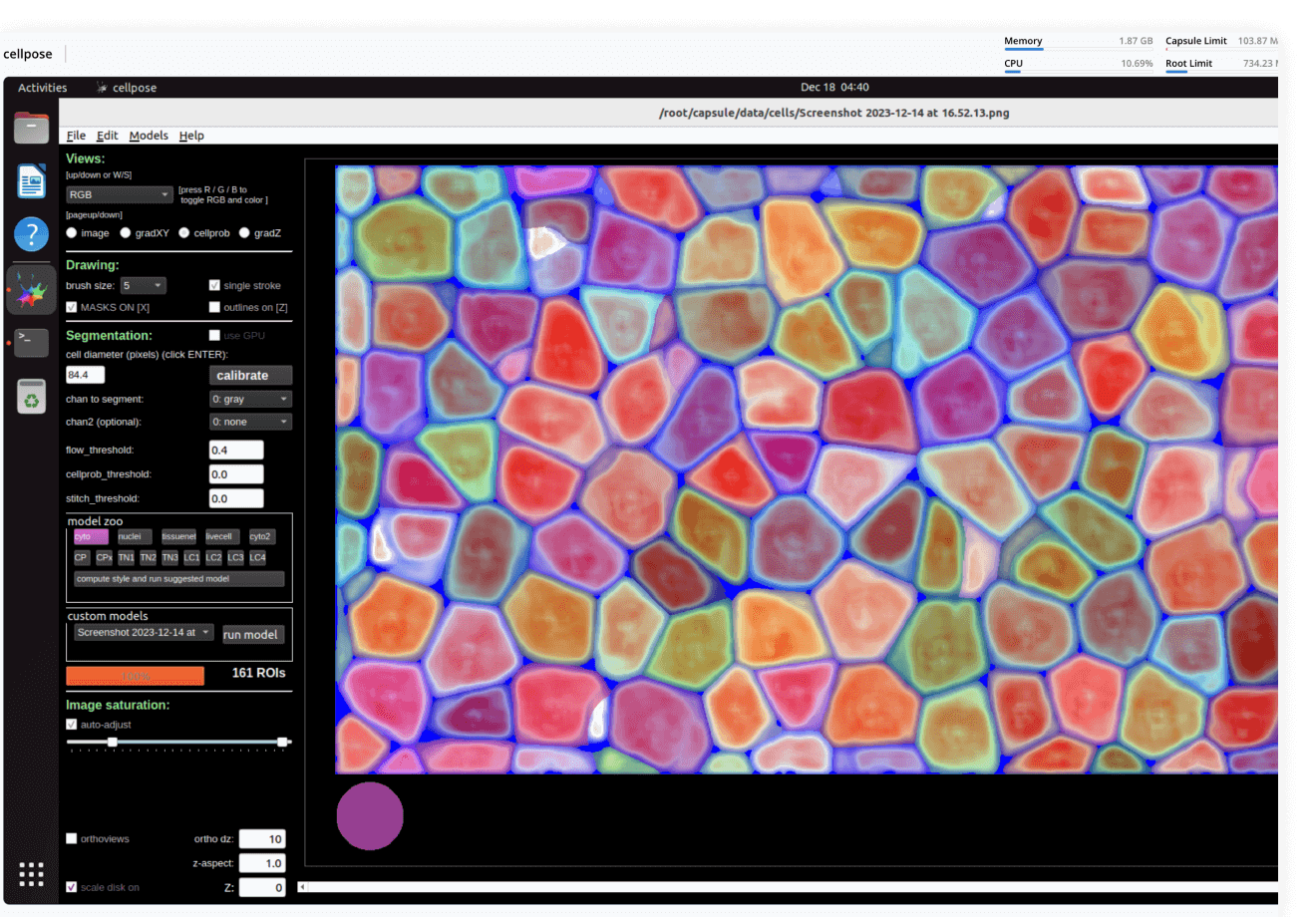

Imaging
Process images using a variety of tools: from dedicated desktop applications to custom-written deep learning pipelines, from a few individual files to petabyte-sized datasets. No DevOps required, always with lineage.
-
Cloud management


Cloud management
Code Ocean makes it easy to manage data and provision compute: CPUs, GPUs, and RAM. Assign flex machines and dedicated machines to manage what is available to your users. Spot instances, idleness detection, and automated shutdown help reduce cloud costs.
-
Data/model provenance

Data/model provenance
Keep track of all data and results with automated result provenance and lineage graph generation. Assess reproducibility with a visual representation of every Capsule, Pipeline, and Data asset involved in a computation.

Data analysis
Use ready-made template Compute Capsules to analyze your data, develop your data analysis workflow in your preferred language and IDE using any open-source software, and take advantage of built-in containerization to guarantee reproducibility.

Data management
Manage your organization's data and control who has access to it. Built specifically to meet all FAIR principles, data management in Code Ocean uses custom metadata and controlled vocabularies to ensure consistency and improve searchability.

Bioinformatics pipelines
Build, configure and monitor bioinformatics pipelines from scratch using a visual builder for easy set-up. Or, import from nf-core in one click for instant access to a curated set of best practice analysis pipelines. Runs on AWS Batch out-of-the-box, so your pipelines scale automatically. No setup needed.

ML model development
Code Ocean is uniquely suited for Artificial Intelligence, Machine Learning, Deep Learning, and Generative AI. Install GPU-ready environments and provision GPU resources in a few clicks. Integration with MLFlow allows you to develop models, track parameters, manage models from development to production, while enjoying out-of-the-box reproducibility and lineage.

Multiomics
Analyze and work with large multimodal datasets efficiently using scalable compute and storage resources, cached packages for R and Python, preloaded multiomics analysis software that works out of the box and full lineage and reproducibility.

Imaging
Process images using a variety of tools: from dedicated desktop applications to custom-written deep learning pipelines, from a few individual files to petabyte-sized datasets. No DevOps required, always with lineage.

Cloud management
Code Ocean makes it easy to manage data and provision compute: CPUs, GPUs, and RAM. Assign flex machines and dedicated machines to manage what is available to your users. Spot instances, idleness detection, and automated shutdown help reduce cloud costs.

Data/model provenance
Keep track of all data and results with automated result provenance and lineage graph generation. Assess reproducibility with a visual representation of every Capsule, Pipeline, and Data asset involved in a computation.


.png?length=720&name=September%20Webinar%20-%20On%20Demand%20(1).png)
